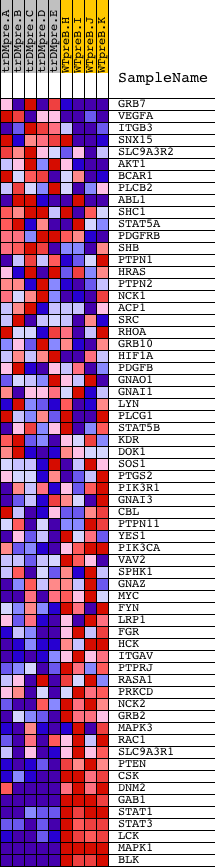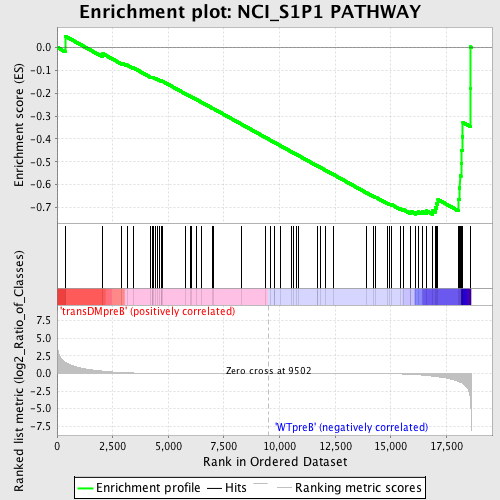
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | NCI_S1P1 PATHWAY |
| Enrichment Score (ES) | -0.73001826 |
| Normalized Enrichment Score (NES) | -1.5632681 |
| Nominal p-value | 0.0043763677 |
| FDR q-value | 0.11406248 |
| FWER p-Value | 0.714 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GRB7 | 20673 | 365 | 1.552 | 0.0488 | No | ||
| 2 | VEGFA | 22969 | 2056 | 0.325 | -0.0279 | No | ||
| 3 | ITGB3 | 20631 | 2915 | 0.120 | -0.0688 | No | ||
| 4 | SNX15 | 23993 3761 | 3142 | 0.089 | -0.0771 | No | ||
| 5 | SLC9A3R2 | 23089 1544 1514 | 3431 | 0.064 | -0.0898 | No | ||
| 6 | AKT1 | 8568 | 4202 | 0.032 | -0.1299 | No | ||
| 7 | BCAR1 | 18741 | 4267 | 0.031 | -0.1320 | No | ||
| 8 | PLCB2 | 5262 | 4304 | 0.030 | -0.1327 | No | ||
| 9 | ABL1 | 2693 4301 2794 | 4331 | 0.029 | -0.1328 | No | ||
| 10 | SHC1 | 9813 9812 5430 | 4437 | 0.027 | -0.1372 | No | ||
| 11 | STAT5A | 20664 | 4526 | 0.025 | -0.1409 | No | ||
| 12 | PDGFRB | 23539 | 4594 | 0.024 | -0.1434 | No | ||
| 13 | SHB | 10493 | 4678 | 0.023 | -0.1469 | No | ||
| 14 | PTPN1 | 5325 | 4699 | 0.023 | -0.1470 | No | ||
| 15 | HRAS | 4868 | 4722 | 0.022 | -0.1472 | No | ||
| 16 | PTPN2 | 23414 | 5771 | 0.012 | -0.2031 | No | ||
| 17 | NCK1 | 9447 5152 | 6001 | 0.011 | -0.2150 | No | ||
| 18 | ACP1 | 4317 | 6044 | 0.011 | -0.2168 | No | ||
| 19 | SRC | 5507 | 6276 | 0.009 | -0.2288 | No | ||
| 20 | RHOA | 8624 4409 4410 | 6499 | 0.009 | -0.2404 | No | ||
| 21 | GRB10 | 4799 | 6979 | 0.007 | -0.2659 | No | ||
| 22 | HIF1A | 4850 | 7024 | 0.007 | -0.2680 | No | ||
| 23 | PDGFB | 22199 2311 | 8288 | 0.003 | -0.3359 | No | ||
| 24 | GNAO1 | 4785 3829 | 9360 | 0.000 | -0.3936 | No | ||
| 25 | GNAI1 | 9024 | 9570 | -0.000 | -0.4049 | No | ||
| 26 | LYN | 16281 | 9749 | -0.001 | -0.4145 | No | ||
| 27 | PLCG1 | 14753 | 9774 | -0.001 | -0.4157 | No | ||
| 28 | STAT5B | 20222 | 10036 | -0.001 | -0.4297 | No | ||
| 29 | KDR | 16509 | 10530 | -0.003 | -0.4562 | No | ||
| 30 | DOK1 | 17104 1018 1177 | 10626 | -0.003 | -0.4612 | No | ||
| 31 | SOS1 | 5476 | 10751 | -0.003 | -0.4677 | No | ||
| 32 | PTGS2 | 5317 9655 | 10832 | -0.003 | -0.4719 | No | ||
| 33 | PIK3R1 | 3170 | 11690 | -0.006 | -0.5178 | No | ||
| 34 | GNAI3 | 15198 | 11718 | -0.006 | -0.5190 | No | ||
| 35 | CBL | 19154 | 11840 | -0.006 | -0.5252 | No | ||
| 36 | PTPN11 | 5326 16391 9660 | 12084 | -0.007 | -0.5380 | No | ||
| 37 | YES1 | 5930 | 12438 | -0.009 | -0.5567 | No | ||
| 38 | PIK3CA | 9562 | 13925 | -0.021 | -0.6358 | No | ||
| 39 | VAV2 | 5848 2670 | 14223 | -0.025 | -0.6507 | No | ||
| 40 | SPHK1 | 5484 1266 1402 | 14313 | -0.027 | -0.6543 | No | ||
| 41 | GNAZ | 71 | 14862 | -0.041 | -0.6820 | No | ||
| 42 | MYC | 22465 9435 | 14956 | -0.045 | -0.6851 | No | ||
| 43 | FYN | 3375 3395 20052 | 15046 | -0.049 | -0.6877 | No | ||
| 44 | LRP1 | 9284 | 15413 | -0.071 | -0.7043 | No | ||
| 45 | FGR | 4723 | 15575 | -0.084 | -0.7093 | No | ||
| 46 | HCK | 14787 | 15902 | -0.120 | -0.7215 | Yes | ||
| 47 | ITGAV | 4931 | 15903 | -0.120 | -0.7162 | Yes | ||
| 48 | PTPRJ | 9664 | 16129 | -0.169 | -0.7209 | Yes | ||
| 49 | RASA1 | 10174 | 16249 | -0.195 | -0.7187 | Yes | ||
| 50 | PRKCD | 21897 | 16416 | -0.233 | -0.7174 | Yes | ||
| 51 | NCK2 | 9448 | 16599 | -0.287 | -0.7145 | Yes | ||
| 52 | GRB2 | 20149 | 16888 | -0.389 | -0.7128 | Yes | ||
| 53 | MAPK3 | 6458 11170 | 17011 | -0.437 | -0.7001 | Yes | ||
| 54 | RAC1 | 16302 | 17052 | -0.451 | -0.6824 | Yes | ||
| 55 | SLC9A3R1 | 11250 | 17116 | -0.476 | -0.6647 | Yes | ||
| 56 | PTEN | 5305 | 18041 | -1.135 | -0.6644 | Yes | ||
| 57 | CSK | 8805 | 18077 | -1.181 | -0.6141 | Yes | ||
| 58 | DNM2 | 4635 3103 | 18109 | -1.237 | -0.5612 | Yes | ||
| 59 | GAB1 | 18828 | 18157 | -1.275 | -0.5074 | Yes | ||
| 60 | STAT1 | 3936 5524 | 18185 | -1.330 | -0.4502 | Yes | ||
| 61 | STAT3 | 5525 9906 | 18210 | -1.378 | -0.3906 | Yes | ||
| 62 | LCK | 15746 | 18240 | -1.451 | -0.3281 | Yes | ||
| 63 | MAPK1 | 1642 11167 | 18579 | -3.793 | -0.1789 | Yes | ||
| 64 | BLK | 3205 21791 | 18586 | -4.095 | 0.0016 | Yes |